McGover Institute for Brain Research at MIT and Broad Institute M M M M M M MM and Harvard .No has re-engineered a compact RNA-way enzyme found in bacteria in an efficient, programmable editor of human DNA.
The proteins they created, called the NovSCB, may be convenient to make certain changes to the genetic code, modulate the activity of specific genes, or to perform other editing tasks. Because its smaller size facilitates delivery in cells, noviscub developers say it is a promising candidate for developing gene therapy to prevent or prevent disease.
The study was led by James of Neuroscience of MIT and Patricia Poitrus Professor Fang Zhang, which is also a key member of the McGover Institute and the investigator of the Howard Hughes Medical Institute and the Broad Institute. Zhang and his team reported their open Access Case work in the journal this month Nature Biotecnovy Ology.
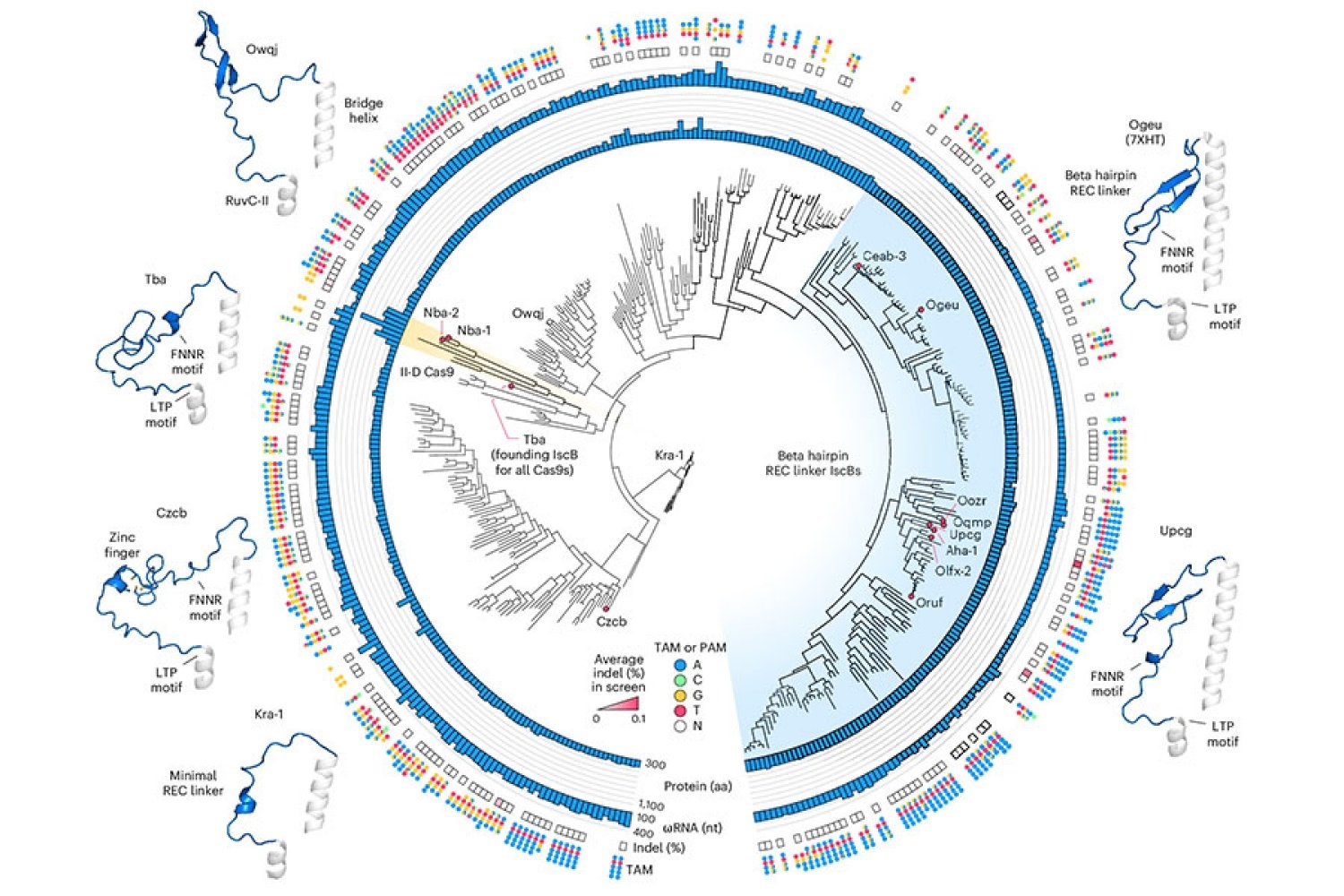
The NovSCB is taken from a bacterial DNA cutter that belongs to a family of protein called ISCBS, which was found in Jhang’s lab in 2021. ISCBS is a type of omega system, CAS9 is the ancestor’s ancestor, part of the bacterial CRISPR system that has developed in Zhang and others in powerful genome-editing tools. Like CAS9, the ISCB enzymes cut DNA on sites specified by the RNA guide. By re -programing that guide, researchers can redirect enzymes to target sequences of their choice.
The ISCBA team’s attention was not taken as they share the main characteristics of CRISPR’s DNA-cutting CAS9, but also because it is a third of its size. It would be an advantage for potential gene therapy: compact tools are easier to deliver to cells, and with small enzymes, researchers were more relieved for tinker, adding potential new functions without creating a very large tool for clinical use.
With his preliminary study of ISCB, researchers from Zhang’s Lab knew that some members of the family could cut DNA goals in human cells. No bacterial protein has worked enough to deploy therapeutically, however: the team will have to change the ISCB to ensure that the team can effectively edit targets in human cells without disturbing the rest of the genome.
To begin that engineering process, Soumya Kannan, a graduate student at Zhang’s lab, is now a junior fellow at Harvard Society FE Fellow, and Postdoc Shyou Zoo first invented an ISCB that will make a good start. They tested about 400 different ISCB enzymes that can be found in bacteria. Were able to edit DNA in ten human cells.
It will also need to enhance the most active of it to make it a useful genome editing tool. The challenge will increase the activity of the enzyme, but only on the sequences mentioned by its RNA guide. If the enzyme becomes more active, but indirectly, it will cut DNA in unnecessary places. Zoo explains, “The key to adjusting the improvement of both activity and uniqueness at the same time.”
Zoo notes that bacterial ISCB is directed by relatively short RNA guides towards their target sequences, making it difficult to limit the activity of the enzyme to a certain part of the genome. If an ISCB can be made an engineer to accommodate a long guide, it is less likely to work on sequences from the front target.
To pt Optimize ISCB for Human Genome Editing, the team took advantage of the information that the graduate student, Han Alte-Tran, who is now postdock at Washington Shington University, learned about the variety of bacterial ISCBs and how they developed. For example, researchers have noted that ISCB, which works in human cells, includes a segment called REC, which was absent in other ISCBs. They suspect that the enzyme may require that segment to communicate with DNA in human cells. When they look closely at the area, structural modeling suggests that by expanding a small portion of the protein, the REC may enable ISCB to identify RNA guides for a long time.
Based on these observations, the team experimented with exchanging parts of the REC domains from various ISCB and CAS9, evaluating how protein function was affected by each change. Guide by their understanding of how both ISCBS and CAS9 interact with DNA and their RNA guides, researchers made additional changes aiming to make both efficiency and uniqueness best.
Finally, they produced a protein, called a noviscub, which was 100 times more active in human cells than the ISCB he started, and showed good uniqueness for its goals.
Kannan and Zoo created and screened hundreds of new ISCBs before reaching the NoviSCB – and every change on the original protein was strategic. Their efforts were guided by the Junowledge of the Natural Evolution of their team’s ISCBS, as well as predictions of how each change would affect the formation of protein, using an artificial intelligence tool called Alfafold 2. Compared to the traditional methods of screening for the introduction of random changes in the protein and, with this rational engineering approach, the ability to identify the protein with the features they were looking for greatly accelerated.
The team showed that NovSCB is a good scaffolding for various genome editing equipment. “It works the same biochemically the same as CAS9, and it makes it easy to bind to Optim ptimized tools with CAS9 scpholds,” says Kannan, “says Kannan. With different changes, researchers used NovSCB to replace the specific characters of the DNA code in human cells and to change the activity of targeted genes.
Importantly, NovSCB-based tools are sufficiently compact to easily pack within a single adeno-related virus (AAV)-vector-vector is commonly used to deliver gene therapy to patients. Because they are bulkier, the equipment developed using KS9 may require a more complex delivery strategy.
Demonstrating the possibility of a noviscub for therapeutic use, Zhang’s team created a tool called Omegaoff that would add chemical markers to DNA to dial the activity of certain genes. They programmed the omegaoff to suppress the gene associated with cholesterol regulation, then the use of AAVs to deliver the living people of the mice, leading to a permanent reduction in the level of cholesterol in the blood of animals.
The team expects that noviscubs can be used to target genome editing tools in most human genes, and look forward to seeing how other labs organize a new technique. They also hope that other people will take their evolutionary-guisted approach to rational protein engineering. “Nature has such a variety, and its systems have different advantages and disadvantages,” says Zu. “By learning about that natural variety, we can create the systems that are trying to engineer better and better.”
This study is partly, K. Lisa Yang and Hawk e. Ten Center for Molecular Therapeutics, MIT, Broad Institute Programable Theraputics Gift Donors, Personing Square Foundation, William Ekman, Nary Oxman, Philips Family, and J. And p. Was funded by Potrus.